Notch signaling: the demise of elegant simplicity | |
|
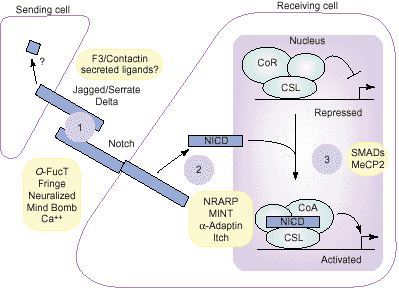
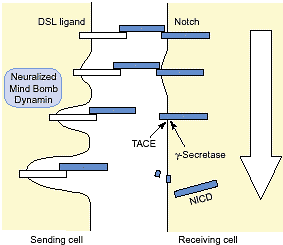
Notch signaling can be viewed as an elegantly simple pathway
that begins when the Notch receptor binds ligand, and ends
when the Notch intracellular domain enters the nucleus and
activates transcription. However, it is becoming increasingly
clear that this core pathway is subject to a wide array of
regulatory influences, from those that affect ligand–receptor
interactions to those that govern the choice of Notch target
genes. Even Notch ligands are now being scrutinized with
respect to the possibility that they, too, function in the nucleus.
A complete understanding of Notch signaling therefore
requires us to look well beyond the core pathway. 1. Artavanis-Tsakonas S, Rand MD, Lake RJ: Notch signaling: cell
fate control and signal integration in development.
Science 1999, 284:770-776.
2. Maillard I, Adler SH, Pear WS: Notch and the immune system.
Immunity 2003, 19:781-791.
3. Weinmaster G, Kintner C: Modulation of Notch Signaling
During Somitogenesis. Annu Rev Cell Dev Biol 2003,
19:367-395.
4. Lai EC: Notch signaling: control of cell communication and
cell fate. Development 2004, 131:965-973.
5. Weng AP, Aster JC: Multiple niches for Notch in cancer: context
is everything. Curr Opin Genet Dev 2004, 14:48-54.
6. Lubman OY, Korolev SV, Kopan R: Anchoring notch genetics
and biochemistry; structural analysis of the ankyrin domain
sheds light on existing data. Mol Cell 2004, 13:619-626.
7. Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR,
Cumano A, Roux P, Black RA, Israel A: A Novel Proteolytic
Cleavage Involved in Notch Signaling: The Role of the
Disintegrin-Metalloprotease TACE. Mol Cell 2000, 5:207-216.
8. Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X,
Pan DJ, Ray WJ, Kopan R: A ligand-induced extracellular
cleavage regulates gamma-secretase-like proteolytic
activation of Notch1. Mol Cell 2000, 5:197-206.
9. Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A:
Signalling downstream of activated mammalian Notch.
Nature 1995, 377:355-358.
10. Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M,
Kintner CR, Evans RM, Kadesch T: A histone deacetylase
corepressor complex regulates the Notch signal transduction
pathway. Genes Dev 1998, 12:2269-2277.
11. Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD: CIR, a
corepressor linking the DNA binding factor CBF1 to the
histone deacetylase complex. Proc Natl Acad Sci USA 1999,
96:23-28.
12. Zhou S, Fujimuro M, Hsieh JJ, Chen L, Miyamoto A, Weinmaster G,
Hayward SD: SKIP, a CBF1-associated protein, interacts with
the ankyrin repeat domain of NotchIC To facilitate NotchIC
function. Mol Cell Biol 2000, 20:2400-2410.
13. Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S,
Griffin JD:MAML1, a human homologue of drosophila
mastermind, is a transcriptional co-activator for NOTCH
receptors. Nat Genet 2000, 26:484-489.
14. Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA:
Mastermind mediates chromatin-specific transcription and
turnover of the Notch enhancer complex. Genes Dev 2002,
16:1397-1411.
15. Jeffries S, Robbins DJ, Capobianco AJ: Characterization of a
high-molecular-weight Notch complex in the nucleus of
Notch(ic)-transformed RKE cells and in a human T-cell
leukemia cell line. Mol Cell Biol 2002, 22:3927-3941.
16. Wallberg AE, Pedersen K, Lendahl U, Roeder RG: p300 and PCAF
act cooperatively to mediate transcriptional activation from
chromatin templates by notch intracellular domains in vitro.
Mol Cell Biol 2002, 22:7812-7819.
17. Nam Y, Weng AP, Aster JC, Blacklow SC: Structural
requirements for assembly of the CSL.intracellular
Notch1.Mastermind-like 1 transcriptional activation complex.
J Biol Chem 2003, 278:21232-21239.
18. Bash J, Zong WX, Banga S, Rivera A, Ballard DW, Ron Y,
Gelinas C: Rel/NF-kappaB can trigger the Notch signaling
pathway by inducing the expression of Jagged1, a ligand
for Notch receptors. EMBO J 1999, 18:2803-2811.
19. Sasaki Y, Ishida S, Morimoto I, Yamashita T, Kojima T, Kihara C,
Tanaka T, Imai K, Nakamura Y, Tokino T: The p53 family member
genes are involved in the Notch signal pathway. J Biol Chem
2002, 277:719-724.
20. Lai EC, Deblandre GA, Kintner C, Rubin GM: Drosophila
neuralized is a ubiquitin ligase that promotes the
internalization and degradation of delta. [see comments].
Dev Cell 2001, 1:783-794.
21. Deblandre GA, Lai EC, Kintner C: Xenopus neuralized is a
ubiquitin ligase that interacts with XDelta1 and regulates
Notch signalling. [see comments]. Dev Cell 2001, 1:795-806.
22. Yeh E, Dermer M, Commisso C, Zhou L, McGlade CJ, Boulianne
GL: Neuralized functions as an E3 ubiquitin ligase during
Drosophila development. Curr Biol 2001, 11:1675-1679.
23. Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY,
Lorick K, Wright GJ, Ariza-McNaughton L et al.: Mind bomb is a
ubiquitin ligase that is essential for efficient activation of
Notch signaling by Delta. Dev Cell 2003, 4:67-82.
24. Seugnet L, Simpson P, Haenlin M: Requirement for dynamin
during Notch signaling in Drosophila neurogenesis.
Dev Biol 1997, 192:585-598.
25. Parks AL, Klueg KM, Stout JR, Muskavitch MA: Ligand
endocytosis drives receptor dissociation and activation in the
Notch pathway. Development 2000, 127:1373-1385.
26. Le Borgne R, Schweisguth F: Unequal segregation of Neuralized
biases Notch activation during asymmetric cell division.
Dev Cell 2003, 5:139-148.
27. Haines N, Irvine KD: Glycosylation regulates Notch signalling.
Nat Rev Mol Cell Biol 2003, 4:786-797.
28. Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R,
Wang Y, Stanley P, Irvine KD, Haltiwanger RS et al.: Fringe is a
glycosyltransferase that modifies Notch. Nature 2000,
406:369-375.
29. Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF,
Weinmaster G: Fringe differentially modulates Jagged1 and
Delta1 signalling through Notch1 and Notch2. Nat Cell Biol
2000, 2:515-520.
30. Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P,
Haltiwanger RS: Modification of epidermal growth factor-like
repeats with O-fucose. Molecular cloning and expression
of a novel GDP-fucose protein O-fucosyltransferase.
J Biol Chem 2001, 276:40338-40345.
31. Sasamura T, Sasaki N, Miyashita F, Nakao S, Ishikawa HO, Ito M,
Kitagawa M, Harigaya K, Spana E, Bilder D et al.: neurotic, a novel
maternal neurogenic gene, encodes an O-fucosyltransferase
that is essential for Notch–Delta interactions. Development
2003, 130:4785-4795.
32. Okajima T, Xu A, Irvine KD: Modulation of notch-ligand binding
by protein O-fucosyltransferase 1 and fringe. J Biol Chem 2003,
278:42340-42345.
33. Lei L, Xu A, Panin VM, Irvine KD: An O-fucose site in the ligand
binding domain inhibits Notch activation. Development 2003,
130:6411-6421.
34. Rand MD, Grimm LM, Artavanis-Tsakonas S, Patriub V,
Blacklow SC, Sklar J, Aster JC: Calcium depletion dissociates
and activates heterodimeric notch receptors. Mol Cell Biol
2000, 20:1825-1835.
35.
Raya A, Kawakami Y, Rodriguez-Esteban C, Ibanes M,
Rasskin-Gutman D, Rodriguez-Leon J, Buscher D, Feijo JA,
Izpisua Belmonte JC: Notch activity acts as a sensor for
extracellular calcium during vertebrate left-right
determination. Nature 2004, 427:121-128. The asymmetric distribution of Nodal and other Notch pathway compo-nents were observed during chick gastrulation. The authors went on to develop a mathematical model that allowed them to test specific inter- actions among those components, leading to the prediction that Notch is activated asymmetrically. The H+/K+-ATPase was also distributed asym-metrically and inhibition of this enzyme ablated asymmetric distribution of Nodal, indicating that the H+/K+ -ATPase acted upstream of Notch. The group went on to show that the H+/K+-ATPase generated a gradient of calcium ions and that this gradient was ultimately responsible for the asymmetric activation of Notch. Co-culture experiments confirmed that Notch’s ability to signal in response to Delta is proportional to the concentration of calcium ions. 36. Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, Takeda Y,
Chia W, Sankar N, Ng YK et al.: F3/contactin acts as a
functional ligand for Notch during oligodendrocyte
maturation. Cell 2003, 115:163-175. Here, it is demonstrated that F3/contactin can bind to Notch on oligo-dendrocyte precursor cells and this leads to the generation of NICD. Whereas Notch signaling from Jagged leads to an inhibition of oligoden-drocyte differentiation, signaling from F3/contactin promoted differentia-tion, suggesting the existence of two distinct pathways. In support of this, the group showed that dominant-negative forms of CSL did not block the effects of F3/contactin, and NICD alone could not recapitulate the effects of F3/contactin. 37. Cui XY, Hu QD, Tekaya M, Shimoda Y, Ang BT, Nie DY, Sun L,
Hu WP, Karsak M, Duka T et al.: NB-3/Notch1 pathway via
Deltex1 promotes neural progenitor cell differentiation into
oligodendrocytes. J Biol Chem 2004.
38. Nofziger D, Miyamoto A, Lyons KM, Weinmaster G: Notch
signaling imposes two distinct blocks in the differentiation of
C2C12 myoblasts. Development 1999, 126:1689-1702.
39. Bush G, diSibio G, Miyamoto A, Denault JB, Leduc R,
Weinmaster G: Ligand-induced signaling in the absence of
furin processing of Notch1. Dev Biol 2001, 229:494-502.
40. Qi H, Rand MD, Wu X, Sestan N, Wang W, Rakic P, Xu T,
Artavanis-Tsakonas S: Processing of the notch ligand
delta by the metalloprotease Kuzbanian. Science 1999,
283:91-94.
41. Mishra-Gorur K, Rand MD, Perez-Villamil B, Artavanis-Tsakonas
S: Down-regulation of Delta by proteolytic
processing. J Cell Biol 2002, 159:313-324.
42. Chen N, Greenwald I: The lateral signal for LIN-12/Notch in
C. elegans vulval development comprises redundant secreted
and transmembrane DSL proteins. Dev Cell 2004, 6:183-192. In an attempt to identify the source of the lateral signal that specifies vulval precursor cells, Chen and Greenwald computationally identified several DSL proteins encoded by the C. elegans genome. One of these, encoded by dsl-1, lacks a transmembrane domain, yet meets the genetic criteria of a bona fide Notch ligand. 43. Small D, Kovalenko D, Kacer D, Liaw L, Landriscina M, Di Serio C,
Prudovsky I, Maciag T: Soluble Jagged 1 represses the function
of its transmembrane form to induce the formation of the
Src-dependent chord-like phenotype. J Biol Chem 2001,
276:32022-32030.
44. LaVoie MJ, Selkoe DJ: The Notch ligands, Jagged and Delta,
are sequentially processed by alpha-secretase and presenilin/
gamma-secretase and release signaling fragments.
J Biol Chem 2003, 278:34427-34437. LaVoie and Selkoe show that Delta and Jagged can each be proteolyzed sequentially to generate a C-terminal fragment, in a manner similar to that observed for the Notch receptor. Interestingly, these C-terminal frag-ments can enter the nucleus where they may affect transcription. 45. Bland CE, Kimberly P, Rand MD: Notch-induced proteolysis and
nuclear localization of the Delta ligand. J Biol Chem 2003,
278:13607-13610.
46. Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M,
Takahashi Y, Thinakaran G, Iwatsubo T: The role of presenilin
cofactors in the gamma-secretase complex. Nature 2003,
422:438-441.
47. Hu Y, Fortini ME: Different cofactor activities in gamma-secretase
assembly: evidence for a nicastrin-Aph-1
subcomplex. J Cell Biol 2003, 161:685-690.
48. Kuroda K, Han H, Tani S, Tanigaki K, Tun T, Furukawa T,
Taniguchi Y, Kurooka H, Hamada Y, Toyokuni S et al.: Regulation
of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 2003, 18:301-312. The authors identify MINT in a two-hybrid screen using CSL as bait and show that MINT can inhibit Notch signaling in cultured cells. Disruption of the MINT gene was found to be embryonic lethal, with embryos displaying multiple developmental defects. The role of MINT in hematopoietic development was assessed using fetal liver cells. MINT did not affect the development of either erythroid or myeloid cells in vitro and caused a decrease in the overall numbers of T cells and B cells in recipient mice. However, in the spleen, the ratio of marginal zone B-cells to follicular B-cells was significantly increased in MINT-/- cells. Given the known effect of Notch on these cells, this is the result expected if MINT acts as an inhibitor of Notch signaling in vivo. 49. Barolo S, Stone T, Bang AG, Posakony JW: Default repression
and Notch signaling: Hairless acts as an adaptor to recruit the
corepressors Groucho and dCtBP to Suppressor of Hairless.
Genes Dev 2002, 16:964-976.
50. Lamar E, Deblandre G, Wettstein D, Gawantka V, Pollet N,
Niehrs C, Kintner C: Nrarp is a novel intracellular component
of the Notch signaling pathway. Genes Dev 2001,
15:1885-1899.
51. Krebs LT, Deftos ML, Bevan MJ, Gridley T: The Nrarp gene
encodes an ankyrin-repeat protein that is transcriptionally
regulated by the notch signaling pathway. Dev Biol 2001,
238:110-119.
52. Yun TJ, Bevan MJ: Notch-regulated ankyrin-repeat protein
inhibits notch1 signaling: multiple notch1 signaling
pathways involved in T cell development. J Immunol 2003,
170:5834-5841.
53. Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN: Asymmetric
localization of a mammalian numb homolog during mouse
cortical neurogenesis. Neuron 1996, 17:43-53.
54. Guo M, Jan LY, Jan YN: Control of daughter cell fates during
asymmetric division: interaction of Numb and Notch.
Neuron 1996, 17:27-41.
55. Spana EP, Doe CQ: Numb antagonizes Notch signaling to
specify sibling neuron cell fates. Neuron 1996, 17:21-26.
56. Berdnik D, Torok T, Gonzalez-Gaitan M, Knoblich JA: The
endocytic protein alpha-Adaptin is required for numb-mediated
asymmetric cell division in Drosophila. Dev Cell 2002,
3:221-231.
57. McGill MA, McGlade CJ: Mammalian numb proteins
promote Notch1 receptor ubiquitination and degradation
of the Notch1 intracellular domain. J Biol Chem 2003,
278:23196-23203.
58. Iso T, Chung G, Hamamori Y, Kedes L: HERP1 is a cell
type-specific primary target of Notch. J Biol Chem 2002,
277:6598-6607.
59. Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I,
Deshaies RJ, Kitajewski J: SEL-10 is an inhibitor of notch
signaling that targets notch for ubiquitin-mediated protein
degradation. Mol Cell Biol 2001, 21:7403-7415.
60. Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F,
Kageyama R: Hes1 and Hes5 as notch effectors in mammalian
neuronal differentiation. EMBO J 1999, 18:2196-2207.
61. Jensen J, Pedersen EE, Galante P, Hald J, Heller RS,
Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD:
Control of endodermal endocrine development by Hes-1.
Nat Genet 2000, 24:36-44.
62. Sumazaki R, Shiojiri N, Isoyama S, Masu M, Keino-Masu K,
Osawa M, Nakauchi H, Kageyama R, Matsui A: Conversion of
biliary system to pancreatic tissue in Hes1-deficient mice.
Nat Genet 2004, 36:83-87.
63. Murtaugh LC, Stanger BZ, Kwan KM, Melton DA: Notch signaling
controls multiple steps of pancreatic differentiation.
Proc Natl Acad Sci USA 2003, 100:14920-14925.
64. Sakata Y, Kamei CN, Nakagami H, Bronson R, Liao JK, Chin MT:
Ventricular septal defect and cardiomyopathy in mice lacking
the transcription factor CHF1/Hey2. Proc Natl Acad Sci USA
2002, 99:16197-16202.
65. Donovan J, Kordylewska A, Jan YN, Utset MF: Tetralogy of fallot
and other congenital heart defects in Hey2 mutant mice.
Curr Biol 2002, 12:1605-1610.
66. Gessler M, Knobeloch K, Helisch A, Amann K, Schumacher N,
Rohde E, Fischer A, Leimeister C: Mouse gridlock. No Aortic
Coarctation or Deficiency, but Fatal Cardiac Defects in
Hey2 S/S Mice. Curr Biol 2002, 12:1601-1604.
67. Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M: The
Notch target genes Hey1 and Hey2 are required for embryonic
vascular development. Genes Dev 2004, 18:901-911.
68. Ross DA, Rao PK, Kadesch T: Dual roles for the Notch target
gene Hes-1 in the differentiation of 3T3-L1 preadipocytes.
Mol Cell Biol 2004, 24:3505-3513.
69. Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA:
Instruction of distinct CD4 T helper cell fates by different notch
ligands on antigen-presenting cells. Cell 2004, 117:515-526. This study conclusively demonstrates that Notch signaling promotes the development of TH2 helper T cells, probably through the direct induction of IL-4 gene transcription. They also provide indirect evidence that supports the idea that development of the TH1 and TH2 subsets is induced by Delta and Jagged, respectively. 70.Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H,
Kubo M, Honjo T: Regulation of alphabeta/gammadelta T cell
lineage commitment and peripheral T cell responses by
Notch/RBP-J signaling. Immunity 2004, 20:611-622. Although not a central conclusion of the paper, Tanigaki and colleagues provide evidence that CSL does not appreciably repress Notch target genes in the absence of Notch activation. 71. Stancheva I, Collins AL, Van den Veyver IB, Zoghbi H, Meehan RR:
A mutant form of MeCP2 protein associated with human
Rett syndrome cannot be displaced from methylated DNA
by notch in Xenopus embryos. Mol Cell 2003, 12:425-435. The authors show that the xHairy2a promoter, a direct target of Notch, is also bound by MeCP2. Paradoxically, a mutant form of MeCP2 that cannot bind the co-repressor SMRT is not displaced from the xHairy2a promoter by Notch. This leads to an overall decrease in the activation of this promoter by Notch and demonstrates that promoter architecture can influence the ability of Notch to activate target genes. 72. Dahlqvist C, Blokzijl A, Chapman G, Falk A, Dannaeus K,
Ibanez CF, Lendahl U: Functional Notch signaling is required
for BMP4-induced inhibition of myogenic differentiation.
Development 2003, 130:6089-6099. A molecular explanation is provided for the crosstalk between the Notch and BMP signaling pathways in muscle development. Although Notch signaling can inhibit muscle differentiation on its own, inhibition by BMP requires sub-inhibitory levels of Notch signaling. This appears to be due to the recruitment of activated SMAD proteins by the NICD–CSL complex. 73. Blokzijl A, Dahlqvist C, Reissmann E, Falk A, Moliner A, Lendahl U,Ibanez CF: Cross-talk between the Notch and TGF-beta
signaling pathways mediated by interaction of the Notch
intracellular domain with Smad3. J Cell Biol 2003, 163:723-728.
74. Schroeter EH, Kisslinger JA, Kopan R: Notch-1 signalling
requires ligand-induced proteolytic release of intracellular
domain. Nature 1998, 393:382-386.
75. Struhl G, Adachi A: Nuclear access and action of notch in vivo.
Cell 1998, 93:649-660.
76. Kidd S, Lieber T, Young MW: Ligand-induced cleavage and
regulation of nuclear entry of Notch in Drosophila
melanogaster embryos. Genes Dev 1998, 12:3728-3740.
77. Iso T, Kedes L, Hamamori Y: HES and HERP families: multiple
effectors of the Notch signaling pathway. J Cell Physiol 2003,
194:237-255. Zfp64 participates in Notch signaling and regulates differentiation in mesenchymal cells. | K.Sakamoto, Y. Tammamura, Ken-chci Katsube, A. Yamaguchi J. of Cell Sci., 121. 1613-1623, 2008 |