Mechanisms of transcriptional repression by histone lysine methylation | |
|
|
Во время развития и гистоны и ДНК вносят вклад в спецификацию и поддержание клеточных особенностей. Репрессивные модификации, как полагают, стабилизируют паттерны экспрессии клеточно-специфичных генов, редуцируя вероятность реактивации генов, не связанных с клонами. В обзоре рассматриваются недавние работы по механизмам, лежащими в основе репрессии, обеспечиваемой Polycomb и метилированием H3K9, и описывается функциональное взаимодействие с активирующим метилированием H3K4. Суммируются недавние данные, которые показывают тесное взаимоотношение между плотностью GC в промоторных последовательностях, связываением транскрипционного фактора и противодействующими активностями определенных эпигенетических регуляторов, таких как histone methyltransferases (HMTs) и histone demethylases (HDMs). Затем мы сравнили сигнатуры хроматина, ассоциированные с разными типами результатов траскрипции от стабильной репрессии до очень динамично регулируемых генов, которые строго показали, что взаимодействие разных эпигенетических путей важно для определения специфических типов наследуемого хроматина и ассоциированных транскрипционных состояний. 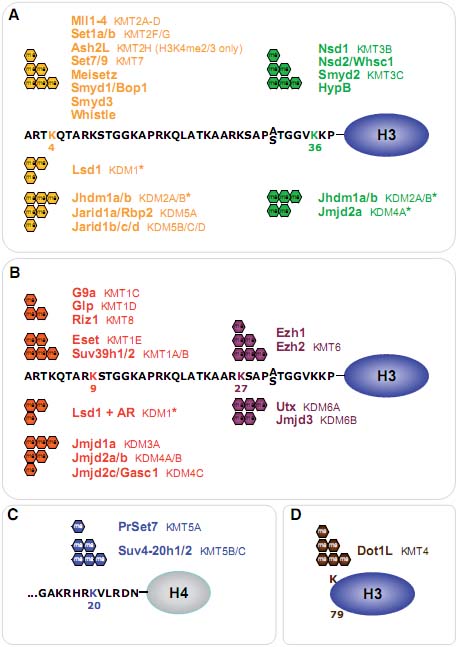 Fig. 1. Overview of histone methylation processes. (A) Methylation of lysines H3K4 and H3K36 is generally correlated with transcriptional activity, and demethylation of H3K4 is required for effective silencing. The specificities of H3K4 and H3K36 HMTs are not unambiguously clear yet. (B) Methylation of H3K9 and H3K27 are hallmarks of transcriptional repression, and the antagonizing HDMs are categorized as transcriptional co-activators. H3K9me3 is a hallmark of constitutive heterochromatin; H3K27me3 is the readout of PcG mediated silencing. (C) In the tail of histone H4, only K20 is targeted by HMTs. H4K20me1 correlates with ongoing transcription, whereas H4K20me3 is an integral part of heterochromatin mediated silencing. (D)Dot1L is the only HMT known to target H3K79 in the globular domain of H3. No HDM is known to target either H4K20 or H3K79. HMTs are indicated above the histone tails, HDMs are below, and hexagons represent the respective methylation status. Asterisks (*) indicate enzymes that target multiple lysine residues.
Fig. 1. Overview of histone methylation processes. (A) Methylation of lysines H3K4 and H3K36 is generally correlated with transcriptional activity, and demethylation of H3K4 is required for effective silencing. The specificities of H3K4 and H3K36 HMTs are not unambiguously clear yet. (B) Methylation of H3K9 and H3K27 are hallmarks of transcriptional repression, and the antagonizing HDMs are categorized as transcriptional co-activators. H3K9me3 is a hallmark of constitutive heterochromatin; H3K27me3 is the readout of PcG mediated silencing. (C) In the tail of histone H4, only K20 is targeted by HMTs. H4K20me1 correlates with ongoing transcription, whereas H4K20me3 is an integral part of heterochromatin mediated silencing. (D)Dot1L is the only HMT known to target H3K79 in the globular domain of H3. No HDM is known to target either H4K20 or H3K79. HMTs are indicated above the histone tails, HDMs are below, and hexagons represent the respective methylation status. Asterisks (*) indicate enzymes that target multiple lysine residues. 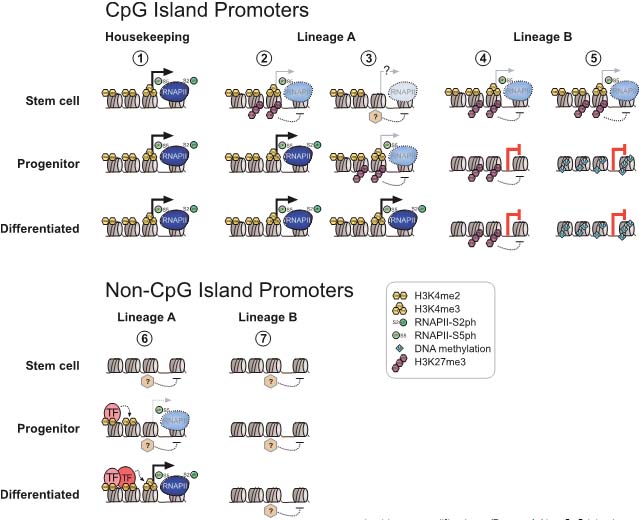 Fig. 2. Models of dynamic chromatin states at CpG island and non-CpG island promoters during differentiation. (Top) CpG-island promoters are H3K4 methylated by default, even when inactive (except when methylated at cytosines). Housekeeping genes are constitutively expressed, marked by H3K4me and elongating RNAPII (1). During differentiation along lineage A, promoters of genes functioning in lineage A harbor a bivalent configuration either at the stem cell or progenitor level and become activated at the subsequent stage (2, 3). In contrast, genes normally expressed in lineage B loose their bivalent configuration during differentiation and remain transcriptionally repressed, either by H3K27me3 or DNA methylation (4, 5). It is currently unclear whether inactive promoters marked by H3K4me2/3 but not H3K27me3 (3) harbor other repressive histone modifications. (Bottom) Non-CpG island promoters require transcription factors (“TF”, 6) to be activated, whereas the mechanisms keeping the repressed state are currently unknown (7). A fraction of genes in lineage A show low levels of H3K4me2 but not H3K4me3 at the progenitor stage, suggesting a transcriptionally poised state. Fig. 2. Models of dynamic chromatin states at CpG island and non-CpG island promoters during differentiation. (Top) CpG-island promoters are H3K4 methylated by default, even when inactive (except when methylated at cytosines). Housekeeping genes are constitutively expressed, marked by H3K4me and elongating RNAPII (1). During differentiation along lineage A, promoters of genes functioning in lineage A harbor a bivalent configuration either at the stem cell or progenitor level and become activated at the subsequent stage (2, 3). In contrast, genes normally expressed in lineage B loose their bivalent configuration during differentiation and remain transcriptionally repressed, either by H3K27me3 or DNA methylation (4, 5). It is currently unclear whether inactive promoters marked by H3K4me2/3 but not H3K27me3 (3) harbor other repressive histone modifications. (Bottom) Non-CpG island promoters require transcription factors (“TF”, 6) to be activated, whereas the mechanisms keeping the repressed state are currently unknown (7). A fraction of genes in lineage A show low levels of H3K4me2 but not H3K4me3 at the progenitor stage, suggesting a transcriptionally poised state.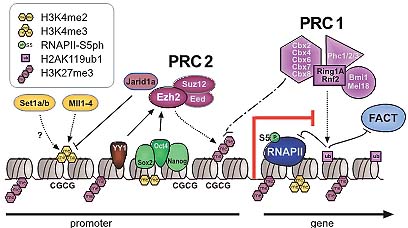 Fig. 3. Transcriptional repression at CpG island bivalent promoters. In mammalian cells, promoters of developmental regulators are marked by both “active” H3K4me2/ 3 and “repressive” H3K27me3, and are therefore termed “bivalent”. Bivalency correlates strongly with high GC density, as present in CpG island promoters. Despite the presence of H3K4me2/3, which is likely mediated by the Mll and/or Set1a/b enzymes, bivalent genes are largely repressed by Polycomb mediated mechanisms. In mammals, targeting of Polycomb complexes is still poorly understood. Of the DNA binding factors recruiting Polycomb in flies, only YY1 is conserved. However, the pluripotency transcription factors Sox2, Oct4 and Nanog co-occupy a large fraction of Polycomb-bound genes, and Oct4 was recently shown to be required for targeting of PRC2 and PRC1 to repressed Oct4 target genes. PRC2-mediated H3K27me3 provides a binding site for PRC1, which in turn mediates monoubiquitination of H2AK119. Moreover, Jarid1a targeted by PRC2 downregulates H3K4me2/3 levels. The initiating form of RNA polymerase II (RNAP-S5P) is present at bivalent genes but is arrested before elongation, presumably by H2AK119ub1 inhibiting recruitment of the remodeling complex FACT. Fig. 3. Transcriptional repression at CpG island bivalent promoters. In mammalian cells, promoters of developmental regulators are marked by both “active” H3K4me2/ 3 and “repressive” H3K27me3, and are therefore termed “bivalent”. Bivalency correlates strongly with high GC density, as present in CpG island promoters. Despite the presence of H3K4me2/3, which is likely mediated by the Mll and/or Set1a/b enzymes, bivalent genes are largely repressed by Polycomb mediated mechanisms. In mammals, targeting of Polycomb complexes is still poorly understood. Of the DNA binding factors recruiting Polycomb in flies, only YY1 is conserved. However, the pluripotency transcription factors Sox2, Oct4 and Nanog co-occupy a large fraction of Polycomb-bound genes, and Oct4 was recently shown to be required for targeting of PRC2 and PRC1 to repressed Oct4 target genes. PRC2-mediated H3K27me3 provides a binding site for PRC1, which in turn mediates monoubiquitination of H2AK119. Moreover, Jarid1a targeted by PRC2 downregulates H3K4me2/3 levels. The initiating form of RNA polymerase II (RNAP-S5P) is present at bivalent genes but is arrested before elongation, presumably by H2AK119ub1 inhibiting recruitment of the remodeling complex FACT.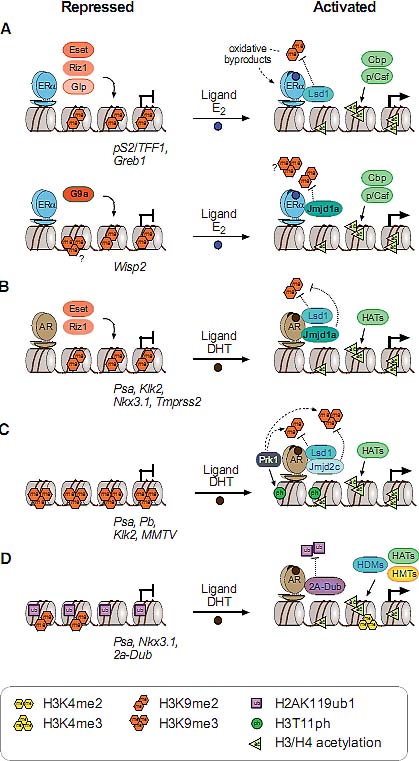 Fig. 4. Nuclear hormone receptors and histone modification pathways. (A) The unliganded Estrogen Receptor (ER) recruits H3K9 HMTs Eset, Glp or Riz1 to target genes that need to be maintained in a repressed state. Upon ligand binding, the HDM Lsd1 clears promoter associated H3K9me2. An alternative ER pathway at distinct target genes involves recruitment of G9a and Jmjd1a. It is not known, how eventual G9a-mediated H3K9me3 is resolved in this scenario (Metivier et al., 2003; Garcia Bassetts et al., 2007; Perillo et al., 2008). (B) Unliganded Androgen Receptor (AR) follows a similar strategy as the ER in the establishment of the preventive “Gatekeeper” situation. However, Glp is not recruited for maintenance of repression, and for the reversal of the silenced state, either Lsd1 or Jmjd1a alone or both enzymes simultaneously are recruited to target promoters (Metzger et al., 2005; Zamane et al., 2006; Garcia Bassetts et al., 2007). (C) The AR can also overcome repression imposed by H3K9me3 by joined recruitment of Jmjd2c and Lsd1 that subsequently transform H3K9me3 into the unmethylated state. H3T11-phosphorylation by Prk1 facilitates removal of the trimethylated form (Metzger et al., 2005, Wissmann et al., 2007; Metzger et al., 2008). (D) The deubiquitinating enzyme 2A-Dub is targeted to AR responsive genes and in concert with H3K9 HDMs and H3K4 HMTs establishes an open chromatin configuration (Zhu et al., 2007; Zhou et al., 2008). Solid arrows indicate enzymatic actions adding PTMs, dotted arrows symbolize removal of the indicated groups, and dashed arrows demonstrate synergistic pathways. Fig. 4. Nuclear hormone receptors and histone modification pathways. (A) The unliganded Estrogen Receptor (ER) recruits H3K9 HMTs Eset, Glp or Riz1 to target genes that need to be maintained in a repressed state. Upon ligand binding, the HDM Lsd1 clears promoter associated H3K9me2. An alternative ER pathway at distinct target genes involves recruitment of G9a and Jmjd1a. It is not known, how eventual G9a-mediated H3K9me3 is resolved in this scenario (Metivier et al., 2003; Garcia Bassetts et al., 2007; Perillo et al., 2008). (B) Unliganded Androgen Receptor (AR) follows a similar strategy as the ER in the establishment of the preventive “Gatekeeper” situation. However, Glp is not recruited for maintenance of repression, and for the reversal of the silenced state, either Lsd1 or Jmjd1a alone or both enzymes simultaneously are recruited to target promoters (Metzger et al., 2005; Zamane et al., 2006; Garcia Bassetts et al., 2007). (C) The AR can also overcome repression imposed by H3K9me3 by joined recruitment of Jmjd2c and Lsd1 that subsequently transform H3K9me3 into the unmethylated state. H3T11-phosphorylation by Prk1 facilitates removal of the trimethylated form (Metzger et al., 2005, Wissmann et al., 2007; Metzger et al., 2008). (D) The deubiquitinating enzyme 2A-Dub is targeted to AR responsive genes and in concert with H3K9 HDMs and H3K4 HMTs establishes an open chromatin configuration (Zhu et al., 2007; Zhou et al., 2008). Solid arrows indicate enzymatic actions adding PTMs, dotted arrows symbolize removal of the indicated groups, and dashed arrows demonstrate synergistic pathways.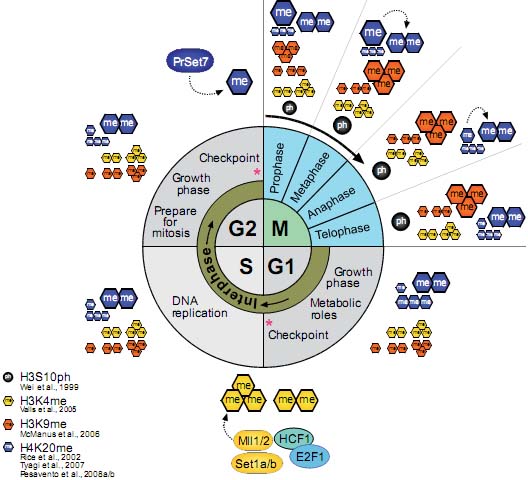 Fig. 5. Histone methylation during the cell division cycle. H3K4me2/3 remains mainly constant during all steps of the cell division cycle. At the G1 to S phase transition, Mll and Set1 complexes are recruited by E2F and HCF1 transcription factors, mediating transcriptional activation of their target genes. H3K9me1/2 remains constant during the cell cycle, only H3K9me3 has a sharp peak at the transition of late G2 to mitosis as revealed by immunofluorescence analyses. H4K20me1 strongly peaks at the G2 to M transition but is rapidly converted to dimethylated H4K20. H4K20me2 levels remain unchanged high at all stages of the cell cycle, and H4K20me3 only slightly peaks at early G1. All H4K20me states were characterized by top down mass spectrometric analyses. The sizes of histone methylation hexagons correspond to their respective levels. Fig. 5. Histone methylation during the cell division cycle. H3K4me2/3 remains mainly constant during all steps of the cell division cycle. At the G1 to S phase transition, Mll and Set1 complexes are recruited by E2F and HCF1 transcription factors, mediating transcriptional activation of their target genes. H3K9me1/2 remains constant during the cell cycle, only H3K9me3 has a sharp peak at the transition of late G2 to mitosis as revealed by immunofluorescence analyses. H4K20me1 strongly peaks at the G2 to M transition but is rapidly converted to dimethylated H4K20. H4K20me2 levels remain unchanged high at all stages of the cell cycle, and H4K20me3 only slightly peaks at early G1. All H4K20me states were characterized by top down mass spectrometric analyses. The sizes of histone methylation hexagons correspond to their respective levels.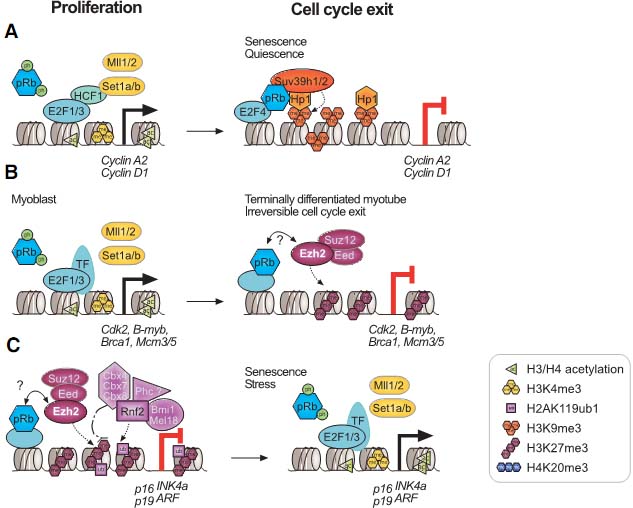 Fig. 6. Suv39h versus Polycomb signaling during the cell division cycle. (A) Cellular senescence or quiescence is mainly maintained by the canonical Suv39h/HP1/Suv4-20h pathway, establishing heterochromatic stretches that are characterized by H3K9me3 and H4K20me3. (B) Terminal differentiation in myoblast cells depends on Ezh2-mediated H3K27me3, while H3K9me3 levels are not altered. Presence of pRb is required, but a direct interaction between pRb and PRC2 members has not been established so far. (C) Cellular proliferation depends on the silencing of the Ink4 locus. pRb recruited Ezh2/H3K27me3 establishes the binding site for the PRC1 complex, that yields H2AK119ub1 and silences the Ink4 locus. Senescence, stress or tumorigenic transformations trigger loss of K27me3 and Ink4 genes are transcribed, resulting in cell cycle exit due to pRb hypo-phosphorylation. Fig. 6. Suv39h versus Polycomb signaling during the cell division cycle. (A) Cellular senescence or quiescence is mainly maintained by the canonical Suv39h/HP1/Suv4-20h pathway, establishing heterochromatic stretches that are characterized by H3K9me3 and H4K20me3. (B) Terminal differentiation in myoblast cells depends on Ezh2-mediated H3K27me3, while H3K9me3 levels are not altered. Presence of pRb is required, but a direct interaction between pRb and PRC2 members has not been established so far. (C) Cellular proliferation depends on the silencing of the Ink4 locus. pRb recruited Ezh2/H3K27me3 establishes the binding site for the PRC1 complex, that yields H2AK119ub1 and silences the Ink4 locus. Senescence, stress or tumorigenic transformations trigger loss of K27me3 and Ink4 genes are transcribed, resulting in cell cycle exit due to pRb hypo-phosphorylation.
Outlook
Recent epigenomic profiling and functional studies have provided insight in the dynamics and regulatory complexity of transcriptional repression, mediated by histone modifying enzymes like HMTs, HDMs, E3 ubiquitin ligases, ubiquitin-specific proteases and other chromatin associated proteins. It is becoming increasingly clear that these machineries function in a sequence dependent manner. Furthermore, the repressed chromatin state is not static but dynamic and reflects the homeostasis between antagonistic enzymatic activities. To truly understand the role of chromatin in transcriptional regulation, it will be necessary to integrate the relative levels of antagonistic histone modifications and their spatial distributions in relation to transcription factor binding sites and RNAPII into the equation. Finally, systematic loss- and gain-of-function experiments are required to dissect the mechanistic hierarchy between the different chromatin and epigenetic modifiers at different stages of development. Beyond doubt, many exciting years are ahead of us.
|